ClusterV-Web
ClusterV-Web is a web application for accurately identifying HIV quasispecies from ONT sequencing data.
ClusterV-Web takes the alignment BAM file, reference FastA file and target region BED file (defining the target regions) of the HIV data as input, and outputs all found quasispecies with their abundance, alignment, variants (SNPs and INDELs), and the drug resistance reports.
Source code are available at this Github page.
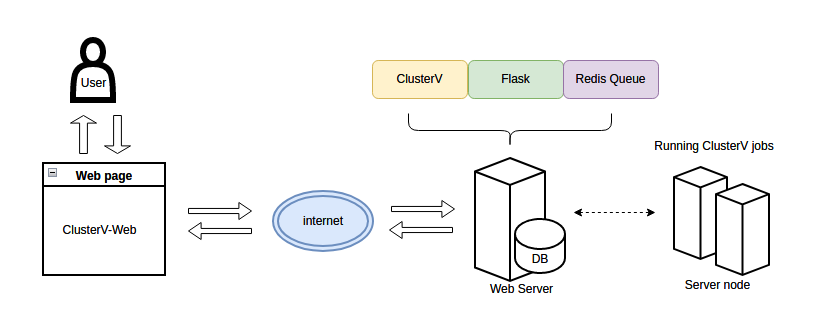
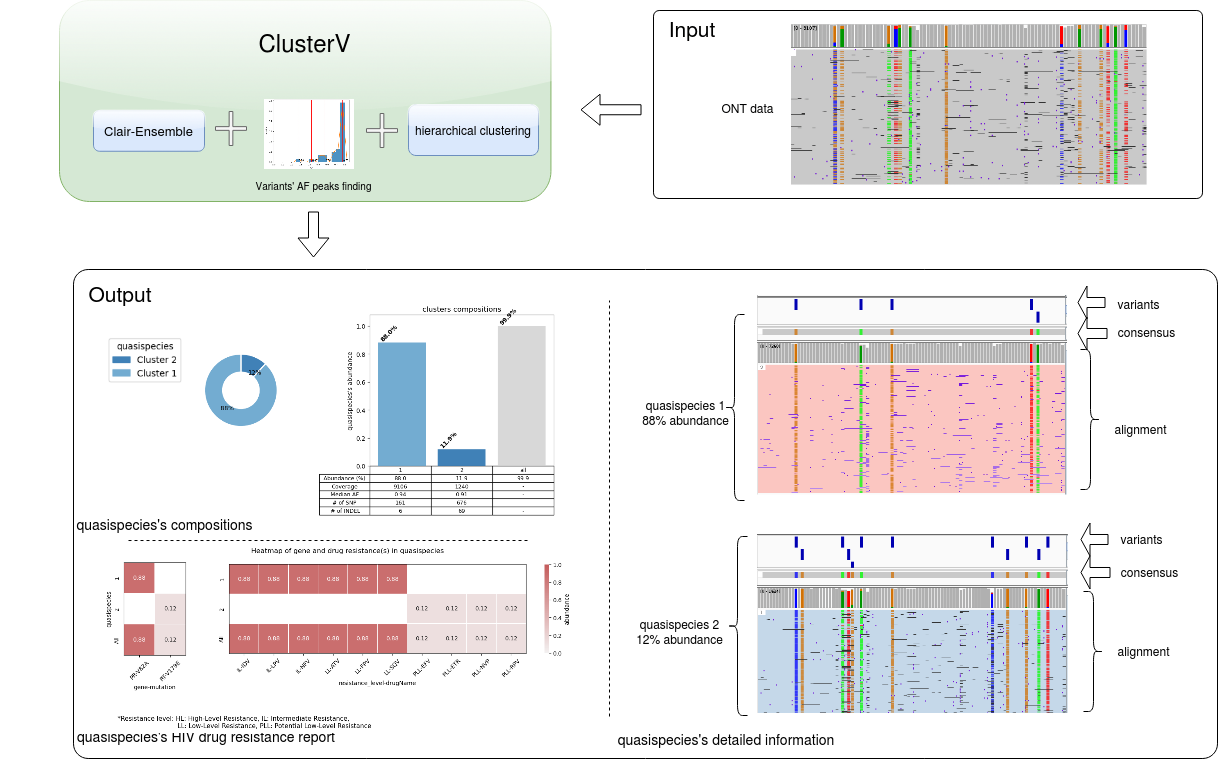
How to use Download example data (BAM ,FASTA and BED)
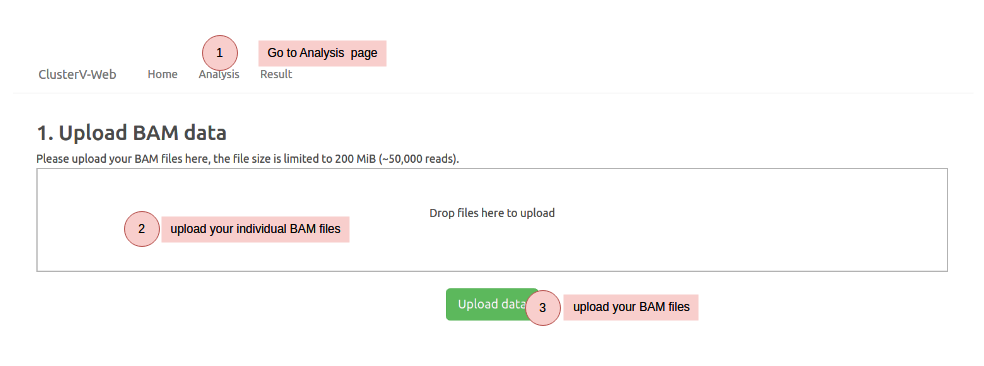
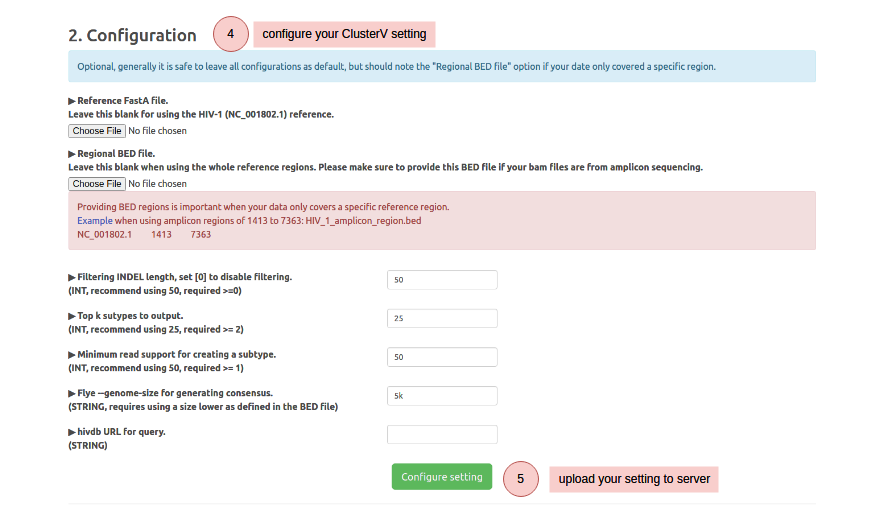
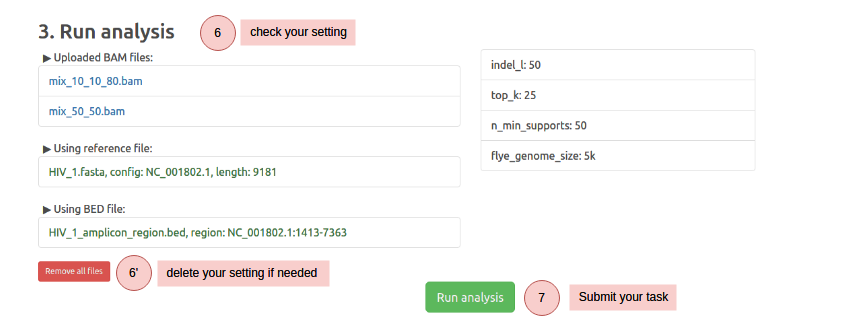
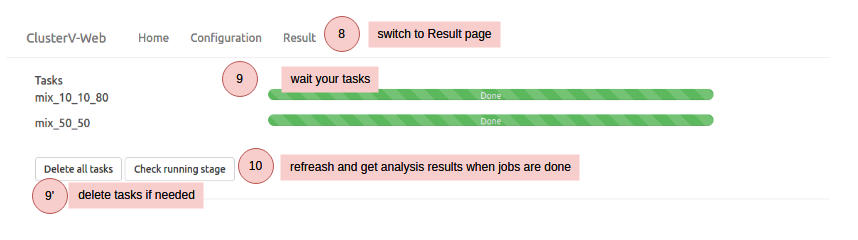
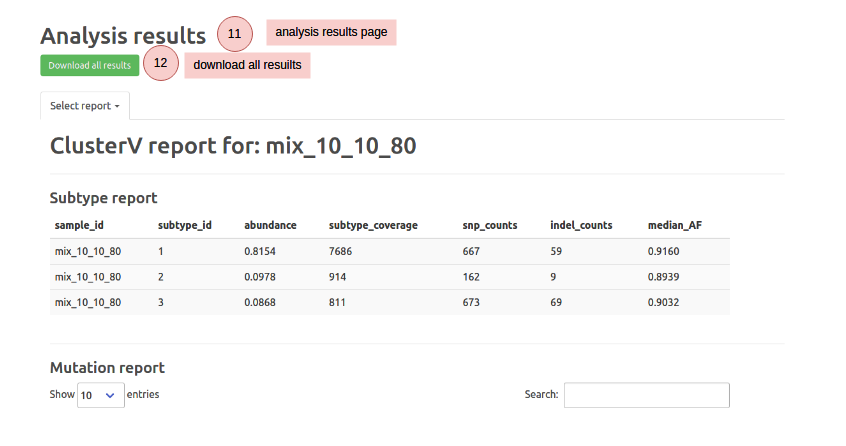
Note that your uploaded data and results will be deleted in one month. We recommend downloading your results for record backup.